Research Interest
I am interested in the application of atomistic and coarse-grained molecular dynamics simulations to investigate protein-ligand interactions. I am also interested in: virtual screening, peptide modification, structural drug design by involving post-translational modifications specifically glycosylation, molecular docking and binding free energy calculations of protein-ligand complexes. I would like to work in close collaboration with other computational chemistry and biology experts to propose testable hypotheses or to test their insights on various drug discovery targets.
Main Projects
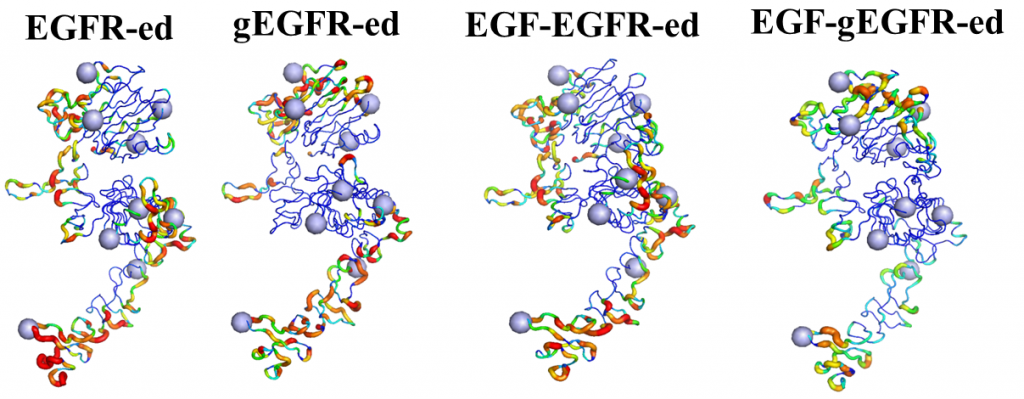
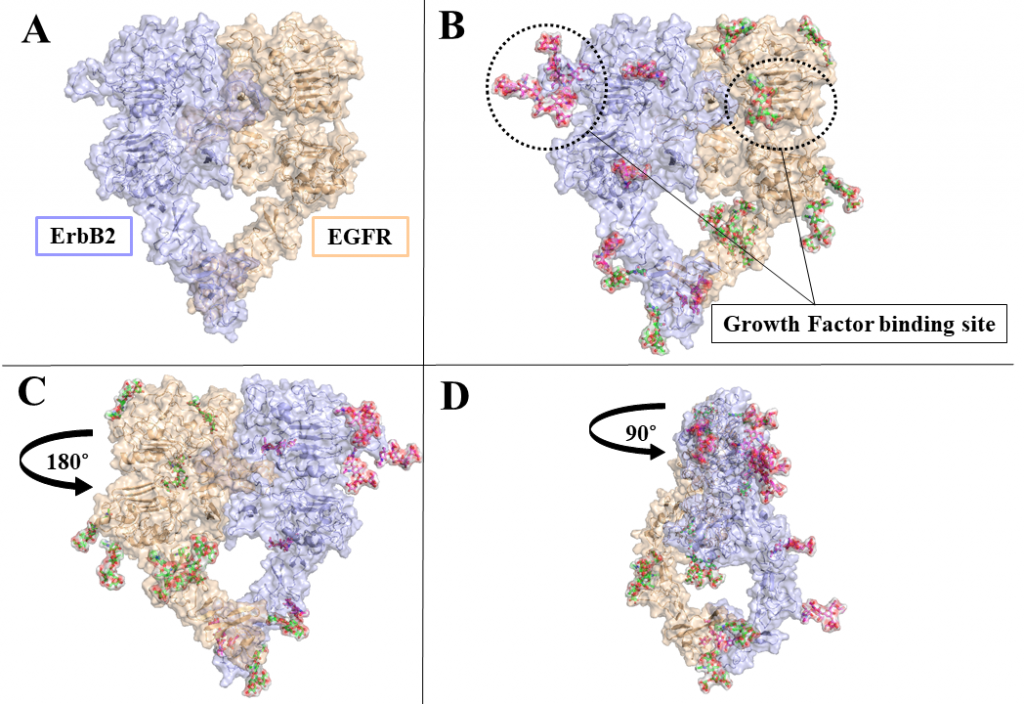
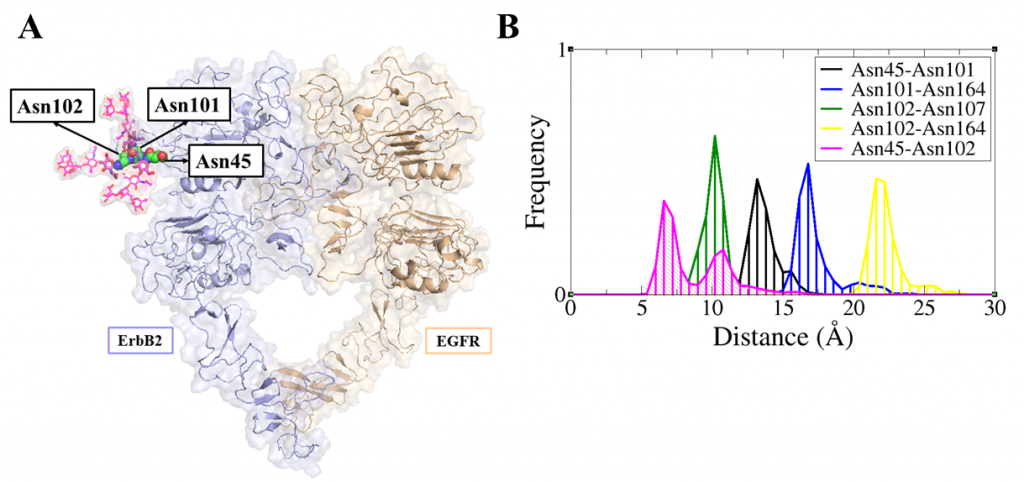
ErbB receptors are tyrosine kinase glycoproteins, overexpressed in several cancers. Their activation triggers various signaling cascades in the cytosol associated with cell proliferation and differentiation. ErbB receptors are activated by binding of growth hormone to their extracellular domain. Activated ErbB receptors form a back to back extended dimer on the cell surface. Various antibodies have been designed to inhibit ErbBs by targeting the growth factor binding site or the dimeric interface. Glycosidically bonded oligosaccharides to ErbBs extracellular domain are known to play a complex role in this activation process. I use molecular dynamics simulations to investigate the role of glycosylation in ErbB receptors functioning.
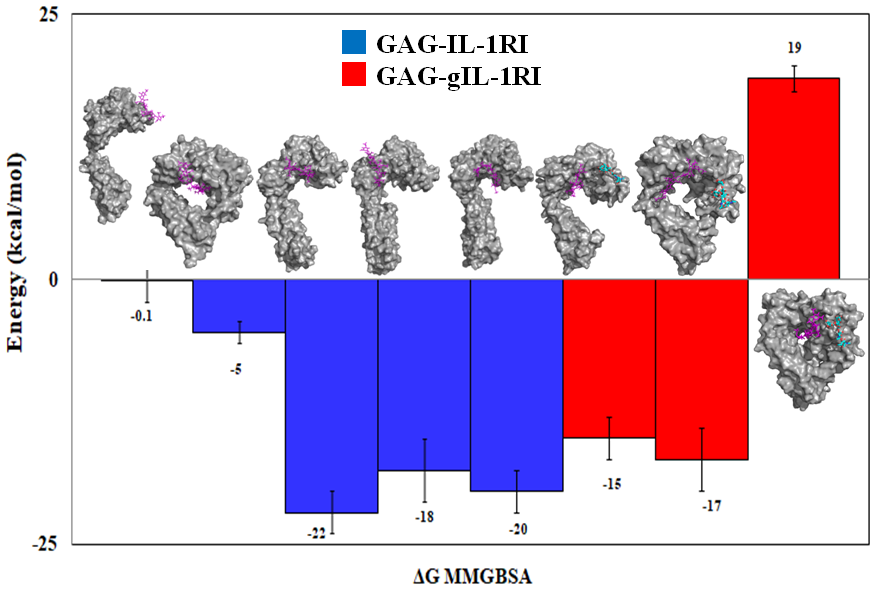
Immune response; IL-1 family of cytokines
IL-1 family of cytokines are key proteins in the regulation of the immune response. IL-1 Receptor Type I (IL-1RI) is their major receptor for triggering the immune responses upon stress and injury. Receptor Type II is an orphan receptor that is homologous to IL-1RI and binds to similar cytokines. I use molecular modelling to study the interplay between these two receptors in different pathological conditions.
SARS-CoV-2 infectivity
Since the COVID-19 outbreak, structural effects of glycosylation in the virion-host cell complex has been among the main subjects of my research work. In a collaborative project, molecular modeling tools were used to show that appearance of a novel glycosylation site on the Receptor Binding Domain of SARS-CoV-2 results in the increased affinity between the virion and the human ACE2 receptor.


N-glycosylated SARS-CoV RBD. The N glycans are distant from the ACE2 binding site.
Side projects
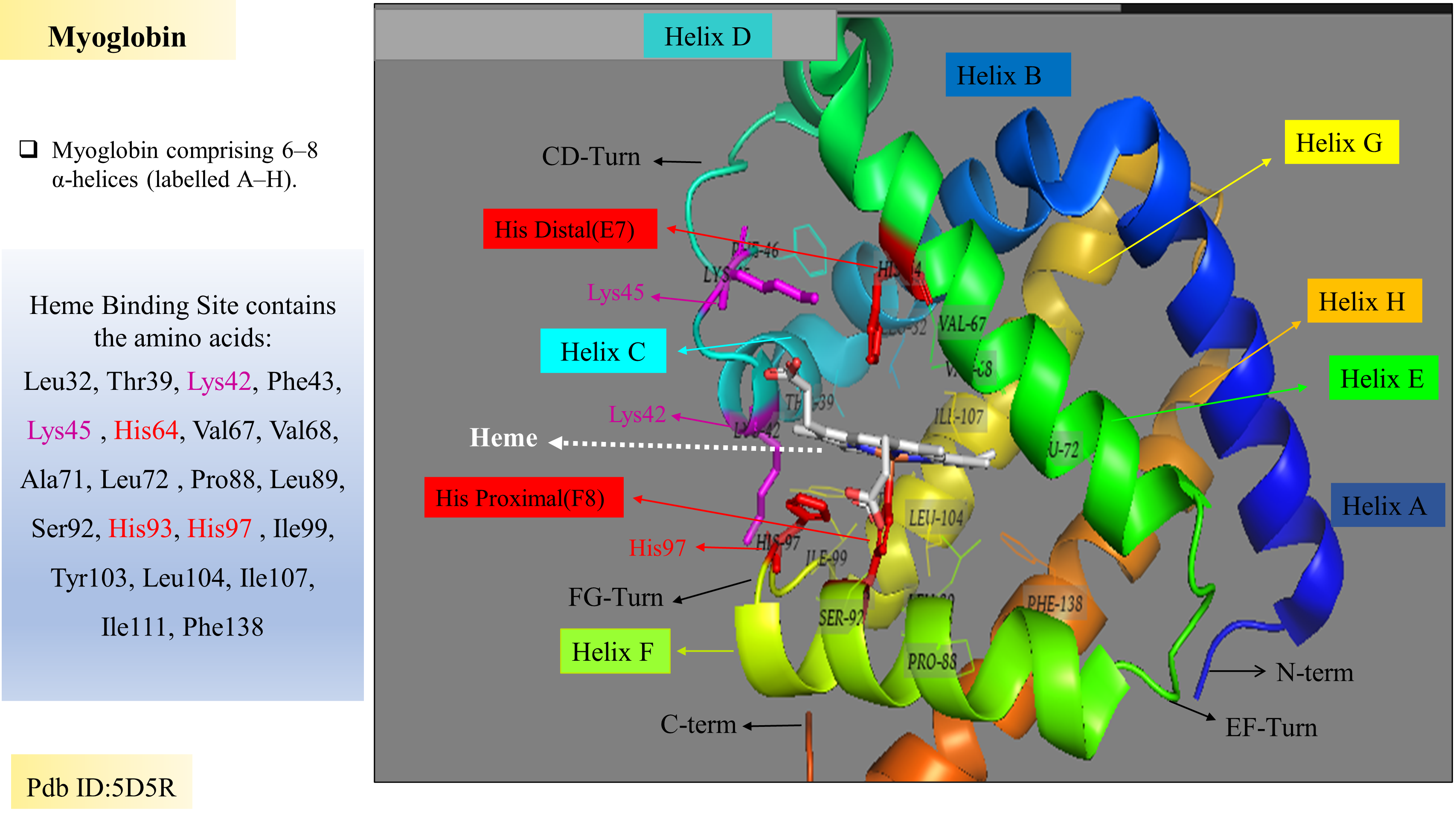
Globins binding sites