 GPMAP
GPMAP GPMAP
GPMAP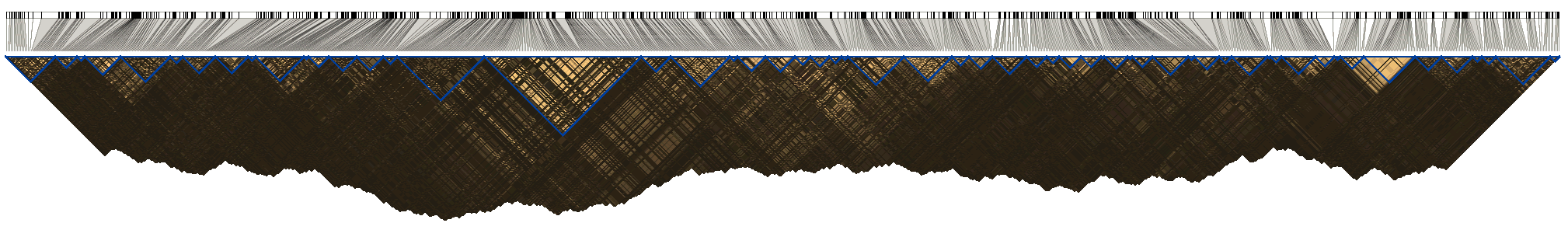
GPMAP divides genome into haplotype blocks (chromosome segments) within which allelic association of SNP pairs are maximized. In genome-wide association studies, bracketing haplotype samples by GPMAP blocks reduces time of computation, though the optimal feature of these blocks guarantees that the highest possible accuracy can be achieved. Besides, the pairwise allelic association between SNPs sketches out patterns of genomic variation in human population, in particular the recombination hotspots. Please cite the following paper if you use this software,
This software has been developed based on the open source software Haploview (ver. 4.3).
GPMAP inherits all Haploview functions, so the user familiar to Haploview should easily find how to run new features of the Haploview+GPMAP package.
The complete source includes number of samples selected from HapMap ENCODE dataset. Although, you can get them directly from here. Note that these samples should be applied as "Haps Format" in the load dialog box at starting the software.
Download the executable JAR file and click on "Haploview.jar". The program will be simply run if the JAR extention is associated with JRE on your machine.
The program is also run by entering this command in console window.
$ java -jar Haploview.jar
| Executable JAR file | [ Haploview+GPMAP ] | |
| Complete source | [ GPMAP.tar.gz ] |
Ali Katanforoush, <a_katanforosh@sbu.ac.ir>, Department of Computer Science, Shahid Beheshti University, G.C., Tehran.
Time-stamp: "2010-09-18 19:47:23 katanforoush"